D Interpretation of Coupling Matrices
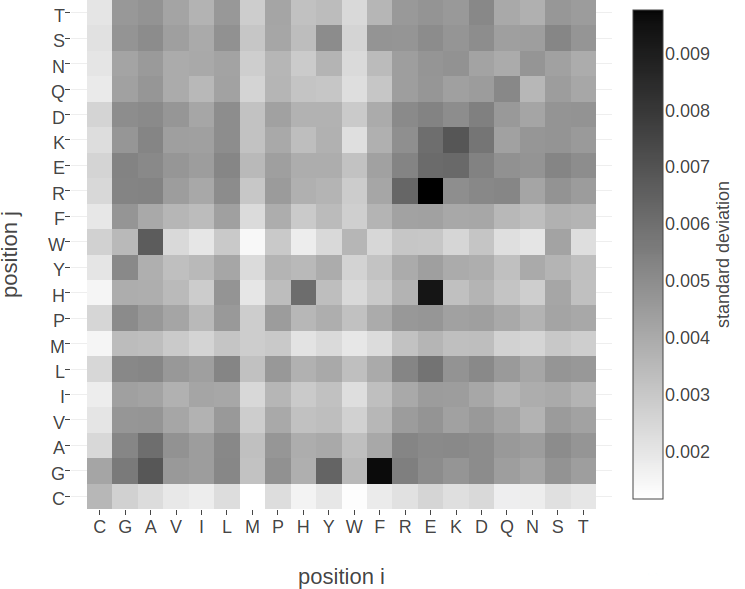
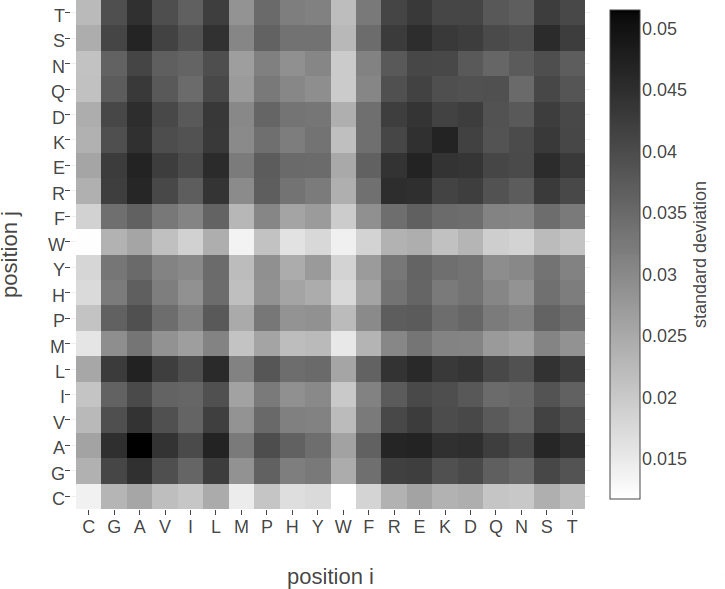
Figure D.1: Standard deviation of squared coupling values \({\wijab}^2\) and of coupling values \(\wijab\) for residue pairs not in physical contact (\(\Delta \Cb > 25 \angstrom\)). Dataset contains 100.000 residue pairs per class (for details see methods section 2.6.6). Amino acids are abbreviated with one-letter code and they are broadly grouped with respect to physico-chemical properties listed in Appendix B Left Standard deviation of squared coupling values \({\wijab}^2\) . Right Standard deviation of coupling values \(\wijab\).